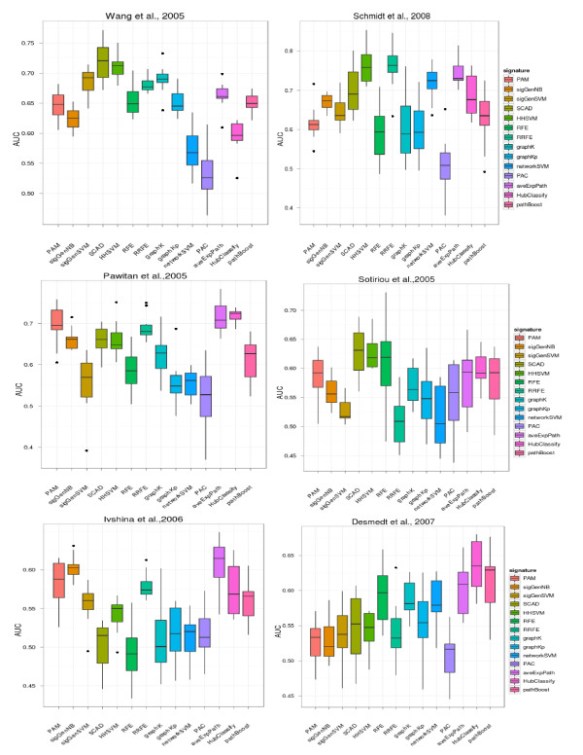
In our recent publication in BMC bioinformatics, we acompared a great deal of feature selection methods to finding prognostic biomakers in 6 breast cancer gene expresion data. No methods show significant performacne in prediction accuracy, feature selection stability and biogical interprety, which against previeous reseach results: current network-based appraoch did not show much benift in our analysis. Meanwhile, A group from NKI also show the simliar results in PloS One. The R codes for these algorithms in our paper is availiable as request.
Category Archives: Computational Genomics
comutational courese in Plos Computational Biology
Short introduction paper in different ares in computational biology.
Fran Lewitter, Welcome to PLoS Computational Biology “Education”
Kenzie D MacIsaac, Ernest Fraenkel, Practical Strategies for Discovering Regulatory DNA Sequence Motifs, April 2006
Duncan Brown, Kimmen Sjölander, Functional Classification Using Phylogenomic Inference,June 2006
Philip E Bourne, Johanna McEntyre, Biocurators: Contributors to the World of Science,October 2006
Yuan Qi, Hui Ge, Modularity and Dynamics of Cellular Networks, December 2006
FrSVM: A filter ranking feature selection algorithm
We use a simple filter feature selection algorithm, called FrSVM, which selected the top ranked genes in PPI network and then training these top raked genes in L2-SVM. FrSVM integrates protein-protein interaction (ppi) network information into feature/gene selection algorithm for prognostic biomarker discovery.
As L2-SVM could not do feature the the ranking of genes were used as feature selection step. Central genes always plays an important role biological process, so make using GeneRank to selected those genes with large differences in their expression.
We applied FrSVM to several cancer datasets and reveals a significantly better prediction performance and higher signature stability. Related manuscript already put to arXiv and R code for FrSVM available at:
Codes: https://sites.google.com/site/yupengcun/software/frsvm
Papers: http://arxiv.org/abs/1212.3214
. Any comments and question on the FrSVM are welcomed. The following is how to run the program:
1. Geting gene expression profiles (GEP), PPi Network.
# Geing GEP
#———————————————————————————-
a = getGEO(“GSExxxxx”, destdir=”/home/YOURPATH/”)
## Normalized the GEP by limma
x= t(normalizeBetweenArrays(exprs(a), method=”quantile”) )
## defien your classes labes, y, as a factor
y= facotr(“Two Class”)
# mapping probest IDs to Entrez IDs
# take hgu133a paltform as example
#———————————————————————————
library(‘hgu133a.db’)
mapped.probes<-mappedkeys(hgu133aENTREZID)
refseq<-as.list(hgu133aENTREZID[mapped.probes])
times<-sapply(refseq, length)
mapping <- data.frame(probesetID=rep(names(refseq),times=times), graphID=unlist(refseq),row.names=NULL, stringsAsFactors=FALSE)
mapping<- unique(mapping)##############################################
# Summarize probests to genes of x by limma
# ad.ppi: Adjacencen matrix of PPI network
Gsub=ad.ppi
mapping <- mapping[mapping[,’probesetID’] %in% colnames(x),]
int <- intersect(rownames(Gsub), mapping[,”graphID”])
xn.m=xn.m[,mapping$probesetID]
index = intersect(mapping[,’probesetID’],colnames(xn.m))
x <- x[,index]
colnames(xn.m) <- map2entrez[index]
ex.sum = t(avereps(t(xn.m), ID=map2entrez[index]))
int= intersect(int, colnames(ex.sum))
ex.sum=ex.sum[,int] ## GEP which matched to PPI network
Gsub=Gsub[int,int] ## PPI network which matched to GEP
2. Run FrSVM program
# You need install for flowing packages for run FrSVM.R programs:
# library(ROCR)
# library(Matrix)
# library(kernlab)
#
## If you want to running parallelly, you also need to load:
# library(multicore)
#
## Here is an expale for 5 times 10-folds Cross-Validtaion
source(“../FrSVM.R”)
res <- frSVM.cv(x=ex.sum, y=y, folds=10,Gsub=Gsub, repeats=5, parallel = FALSE, cores = 2, DEBUG=TRUE,d=0.5,top.uper=0.95,top.lower=0.9)
## the AUC values for 5*10-folds CV
AUC= res$auc